Журнал «Здоровье ребенка» Том 19, №4, 2024
Вернуться к номеру
Аберантне метилювання ДНК, пов’язане з розвитком метаболічно-асоційованої жирової хвороби печінки
Авторы: Абатуров О.Є., Нікуліна А.О., Русакова О.О.
Дніпровський державний медичний університет, м. Дніпро, Україна
Рубрики: Педиатрия/Неонатология
Разделы: Справочник специалиста
Версия для печати
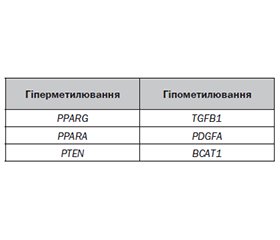
Літературний огляд присвячений висвітленню ключового епігенетичного механізму, що контролює активність транскрипції генів, відіграє вирішальну роль у формуванні геномного імпринтингу, сайленсингу генів, інактивації X-хромосоми, сплайсингу РНК, репарації ДНК, клітинному диференціюванні й перепрограмуванні клітин, а також визначає виникнення, розвиток стеатотичного ураження печінки і метаболічних порушень, — метилювання ДНК. Метилювання цитозиндинуклеотиду (CpG) ДНК буває двох видів: метилювання de novo CpG, яке здійснюють райтери 5mC ДНК — ДНК-метилтрансферази (DNA-(cytosine-5)-methyltransferase — DNMT) 3a і 3b, і підтримуюче метилювання ДНК, що виконує DNMT1 під час реплікації ДНК. Встановлено, що підтримуюче метилювання ДНК дозволяє зберігати в клітинах нової генерації патерн метилювання, характерний для клітин-попередників, а метилювання ДНК тіла гена асоційоване з підвищеною його експресією. Активне деметилювання 5mC здійснюється діоксигеназами ТЕТ, ензиматичними представниками яких є TET1, TET2 і TET3. Продемонстровано, що аберантне метилювання нуклеотидів ДНК безпосередньо пов’язане з активністю синтезу ліпідів, ступенем оксидативного стресу, розвитком стеатозу печінки, низькорівневого запалення, інсулінорезистентності й прогресуванням фіброзу печінки. Автори детально подали функції та особливості ДНК-метилтрансфераз, ластиків і ридерів сайтів 5mC; можливі порушення балансу активності райтерів і ластиків 5mC ДНК; ландшафт і патерни метилювання ДНК; клінічне значення сигнатур метилювання ДНК при метаболічно-асоційованій жировій хворобі печінки. У хворих на метаболічно-асоційовану жирову хворобу печінки спостерігається глобальне гіпометилювання геному — як мінімум 55 генів. Автори наголошують на тому, що використання сигнатур метилювання ДНК є перспективним напрямом ранньої діагностики та прогнозування перебігу метаболічно-асоційованої жирової хвороби печінки, тоді як вивчення молекулярних компонентів механізмів метилювання ДНК, що беруть участь у регуляції експресії генів, залежності їх активності від впливу експосоми дозволить персоніфікувати й удосконалити рекомендації щодо модифікації способу життя та дієти у хворих з метаболічно-асоційованою жировою хворобою печінки.
The literature review deals with DNA methylation, a key epigenetic mechanism that controls the activity of gene transcription, plays a decisive role in the formation of genomic imprinting, gene silencing, X-chromosome inactivation, RNA splicing, DNA repair, cell differentiation and cell reprogramming, and also determines the occurrence and development of liver steatotic lesions and metabolic disorders. Methylation of DNA cytosine dinucleotide (CpG) can be represented in two types: de novo CpG methylation, which is carried out by 5mC DNA writers — DNA-(cytosine-5)-methyltransferase (DNMT) 3a and 3b, and supporting DNA methylation, which is performed by DNMT1 during DNA replication. It has been found that the maintenance DNA methylation allows the preservation of the methylation pattern characteristic of progenitor cells in the cells of the new generation, and the DNA methylation of the gene body is associated with its increased expression. Active demethylation of 5mC is carried out by TET dioxygenases, including three enzymatic representatives: TET1, TET2 and TET3. It has been demonstrated that aberrant methylation of DNA nucleotides is directly related to the activity of lipid synthesis, the degree of oxidative stress, the development of liver steatosis, low-grade inflammation, insulin resistance, and the progression of liver fibrosis. The authors presented in detail the functions and features of DNA methyltransferases, erasers, and readers of 5mC sites; possible violations of the balance of activity of writers and erasers of 5mC DNA; DNA methylation landscape and patterns; clinical significance of DNA methylation signatures in metabolic dysfunction-associated fatty liver disease. Global hypomethylation of genome, at least 55 genes, is observed in patients with metabolic dysfunction-associated fatty liver disease. The authors emphasize that the use of DNA methylation signatures is a promising direction for early diagnosis and prognosis of the course of metabolic dysfunction-associated fatty liver disease, while the study of molecular components of DNA methylation mechanisms involved in the regulation of gene expression, the dependence of their activity on exposure to the exposome will allow to personalize and improve recommendations for lifestyle and diet modification in patients with metabolic dysfunction-associated fatty liver disease.
діти; ожиріння; метаболічно-асоційована жирова хвороба печінки; метилювання ДНК
children; obesity; metabolic dysfunction-associated fatty liver disease; DNA methylation
Для ознакомления с полным содержанием статьи необходимо оформить подписку на журнал.
- Lee J, Kim Y, Friso S, Choi SW. Epigenetics in non-alcoholic fatty liver disease. Mol Aspects Med. 2017 Apr;54:78-88. doi: 10.1016/j.mam.2016.11.008. Epub 2016 Nov 23. PMID: 27889327.
- Sookoian S, Pirola CJ, Valenti L, Davidson NO. Genetic Pathways in Nonalcoholic Fatty Liver Disease: Insights From Systems Biology. Hepatology. 2020 Jul;72(1):330-346. doi: 10.1002/hep.31229. PMID: 32170962; PMCID: PMC7363530.
- Rodríguez-Sanabria JS, Escutia-Gutiérrez R, Rosas-Campos R, Armendáriz-Borunda JS, Sandoval-Rodríguez A. An Update in Epigenetics in Metabolic-Associated Fatty Liver Disease. Front Med (Lausanne). 2022 Jan 11;8:770504. doi: 10.3389/fmed.2021.770504. PMID: 35087844; PMCID: PMC8787199.
- Geiger M, Gorica E, Mohammed SA, Mongelli A, Mengozi A, Delfine V et al. Epigenetic Network in Immunometabolic Disease. Adv Biol (Weinh). 2024 Jan;8(1):e2300211. doi: 10.1002/adbi.202300211. Epub 2023 Oct 4. PMID: 37794610.
- Abaturov A, Nikulina A. Role of genetic modification of the PNPLA3 gene in predicting metabolically unhealthy obesity and associated fatty liver disease in children. Eur J Clin Exp Med. 2023;21(1):5-13. doi: 10.15584/ejcem.2023.1.1.
- Botello-Manilla AE, Chávez-Tapia NC, Uribe M, Nuño-Lámbarri N. Genetics and epigenetics purpose in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2020 Aug;14(8):733-748. doi: 10.1080/17474124.2020.1780915. Epub 2020 Jun 22. PMID: 32552211.
- Aggeletopoulou I, Kalafateli M, Tsounis EP, Triantos C. Epigenetic Regulation in Lean Nonalcoholic Fatty Liver Disease. Int J Mol Sci. 2023 Aug 16;24(16):12864. doi: 10.3390/ijms241612864. PMID: 37629043; PMCID: PMC10454848.
- Fu S, Debes JD, Boonstra A. DNA methylation markers in the detection of hepatocellular carcinoma. Eur J Cancer. 2023 Sep;191:112960. doi: 10.1016/j.ejca.2023.112960. Epub 2023 Jun 28. PMID: 37473464.
- Njei B, Al-Ajlouni YA, Ugwendum D, Abdu M, Forjindam A, Mohamed MF. Genetic and epigenetic determinants of non-alcoholic fatty liver disease (NAFLD) in lean individuals: a systematic review. Transl Gastroenterol Hepatol. 2023 Dec 6;9:11. doi: 10.21037/tgh-23-31. PMID: 38317742; PMCID: PMC10838615.
- Zhang N, Tian X, Yan T, Wang H, Zhang D, Lin C, Liu Q, Jiang S. Insights into the role of nucleotide methylation in metabolic-associated fatty liver disease. Front Immunol. 2023 Mar 20;14:1148722. doi: 10.3389/fimmu.2023.1148722. PMID: 37020540; PMCID: PMC10067741.
- Johnson TB, Coghill RD. The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the Tubercle bacillus. J Amer Chem Society. 1925;547:2838-2844.
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226-32. PMID: 1111098.
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9-25. doi: 10.1159/000130315. PMID: 1093816.
- Horsburgh S, Robson-Ansley P, Adams R, Smith C. Exercise and inflammation-related epigenetic modifications: focus on DNA methylation. Exerc Immunol Rev. 2015;21:26-41. PMID: 25826329.
- Rausch C, Hastert FD, Cardoso MC. DNA Modification Readers and Writers and Their Interplay. J Mol Biol. 2020 Mar 13;432(6):1731-1746. doi: 10.1016/j.jmb.2019.12.018. Epub 2019 Dec 20. PMID: 31866298.
- Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, Cao L. DNA methylation, its mediators and genome integrity. Int J Biol Sci. 2015 Apr 8;11(5):604-17. doi: 10.7150/ijbs.11218. PMID: 25892967; PMCID: PMC4400391.
- Абатуров О.Є., Крючко Т.О., Агафонова О.О. та співавт. Геномний імпринтинг та імпринтинг-асоційовані захворювання. Том 1. Загальні уявлення про геномний імпринтинг та епігенетичні механізми. Харків: Планета-Прінт, 2016. 448 с.
- Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017 Sep;18(9):517-534. doi: 10.1038/nrg.2017.33. Epub 2017 May 30. PMID: 28555658.
- Zhang X, Zhang Y, Wang C, Wang X. TET (Ten-eleven translocation) family proteins: structure, biological functions and applications. Signal Transduct Target Ther. 2023 Aug 11;8(1):297. doi: 10.1038/s41392-023-01537-x. PMID: 37563110; PMCID: PMC10415333.
- López-Moyado IF, Ko M, Hogan PG, Rao A. TET Enzymes in the Immune System: From DNA Demethylation to Immunotherapy, Inflammation, and Cancer. Annu Rev Immunol. 2024 Feb 15. doi: 10.1146/annurev-immunol-080223-044610. Epub ahead of print. PMID: 38360546.
- Kriukienė E, Tomkuvienė M, Klimašauskas S. 5-Hydroxymethylcytosine: the many faces of the sixth base of mammalian DNA. Chem Soc Rev. 2024 Mar 4;53(5):2264-2283. doi: 10.1039/d3cs00858d. PMID: 38205583.
- Farsetti A, Illi B, Gaetano C. How epigenetics impacts on human diseases. Eur J Intern Med. 2023 Aug;114:15-22. doi: 10.1016/j.ejim.2023.05.036. Epub 2023 Jun 3. PMID: 37277249.
- Sergeeva A, Davydova K, Perenkov A, Vedunova M. Mechanisms of human DNA methylation, alteration of methylation patterns in physiological processes and oncology. Gene. 2023 Jul 30;875:147487. doi: 10.1016/j.gene.2023.147487. Epub 2023 May 19. PMID: 37211289.
- Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA et al. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009 Jul;51(1):176-86. doi: 10.1016/j.jhep.2009.03.021. Epub 2009 May 3. PMID: 19450891; PMCID: PMC2773516.
- Cui Y, Ru M, Wang Y, Weng L, Haji RA, Liang H et al. Epigenetic regulation of H3K27me3 in laying hens with fatty liver hemorrhagic syndrome induced by high-energy and low-protein diets. BMC Genomics. 2024 Apr 16;25(1):374. doi: 10.1186/s12864-024-10270-w. PMID: 38627644; PMCID: PMC11022457.
- Page A, Paoli P, Moran Salvador E, White S, French J, Mann J. Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J Hepatol. 2016 Mar;64(3):661-73. doi: 10.1016/j.jhep.2015.11.024. Epub 2015 Nov 26. PMID: 26632634; PMCID: PMC4904781.
- Feng LL, Liu RY, An K, Tang S, Wu J, Yang Q. TET3 as a non-invasive screening tool for the detection of fibrosis in patients with chronic liver disease. Sci Rep. 2023 Apr 19;13(1):6382. doi: 10.1038/s41598-023-33564-7. PMID: 37076545; PMCID: PMC10115894.
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012 May 29;13(7):484-92. doi: 10.1038/nrg3230. PMID: 22641018.
- de Mendoza A. A mammalian DNA methylation landscape. Science. 2023 Aug 11;381(6658):602-603. doi: 10.1126/science.adj4904. Epub 2023 Aug 10. PMID: 37561871.
- Lyall MJ, Thomson JP, Cartier J, Ottaviano R, Kendall TJ, Meehan RR, Drake AJ. Non-alcoholic fatty liver disease (NAFLD) is associated with dynamic changes in DNA hydroxymethylation. Epigenetics. 2020 Jan-Feb;15(1-2):61-71. doi: 10.1080/15592294.2019.1649527. Epub 2019 Aug 7. PMID: 31389294; PMCID: PMC6961686.
- Loyfer N, Magenheim J, Peretz A et al. A DNA methylation atlas of normal human cell types. Nature. 2023 Jan;613(7943):355-364. doi: 10.1038/s41586-022-05580-6. Epub 2023 Jan 4. PMID: 36599988; PMCID: PMC9811898.
- Minton K. Mapping the minutiae of the human methylome. Nat Rev Genet. 2023 Mar;24(3):139. doi: 10.1038/s41576-023-00576-y. PMID: 36646844; PMCID: PMC9841940.
- Xie W, Ma LL, Xu YQ, Wang BH, Li SM. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem Biophys Res Commun. 2019 Oct 8;518(1):120-126. doi: 10.1016/j.bbrc.2019.08.018. Epub 2019 Aug 10. PMID: 31405565.
- Lai Z, Chen J, Ding C, Wong K, Chen X, Pu L, Huang Q et al. Association of Hepatic Global DNA Methylation and Serum One-Carbon Metabolites with Histological Severity in Patients with NAFLD. Obesity (Silver Spring). 2020 Jan;28(1):197-205. doi: 10.1002/oby.22667. Epub 2019 Nov 29. PMID: 31785086.
- Meehan RR, Thomson JP, Lentini A, Nestor CE, Pennings S. DNA methylation as a genomic marker of exposure to chemical and environmental agents. Curr Opin Chem Biol. 2018 Aug;45:48-56. doi: 10.1016/j.cbpa.2018.02.006. Epub 2018 Mar 2. PMID: 29505975.
- Mwinyi J, Boström AE, Pisanu C et al. NAFLD is associated with methylation shifts with relevance for the expression of genes involved in lipoprotein particle composition. Biochim Biophys Acta Mol Cell Biol Lipids. 2017 Mar;1862(3):314-323. doi: 10.1016/j.bbalip.2016.12.005. Epub 2016 Dec 18. PMID: 27993651.
- Ahrens M, Ammerpohl O, von Schönfels W et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013 Aug 6;18(2):296-302. doi: 10.1016/j.cmet.2013.07.004. PMID: 23931760.
- Hyun J, Jung Y. DNA Methylation in Nonalcoholic Fatty Liver Disease. Int J Mol Sci. 2020 Oct 30;21(21):8138. doi: 10.3390/ijms21218138. PMID: 33143364; PMCID: PMC7662478.
- Vachher M, Bansal S, Kumar B, Yadav S, Burman A. Deciphering the role of aberrant DNA methylation in NAFLD and NASH. Heliyon. 2022 Oct 18;8(10):e11119. doi: 10.1016/j.heliyon.2022.e11119. PMID: 36299516; PMCID: PMC9589178.
- Ma J, Nano J, Ding J et al. A Peripheral Blood DNA Methylation Signature of Hepatic Fat Reveals a Potential Causal Pathway for Nonalcoholic Fatty Liver Disease. Diabetes. 2019 May;68(5):1073-1083. doi: 10.2337/DB18-1193. Epub 2019 Apr 1. PMID: 30936141; PMCID: PMC6477898.
- Wu J, Zhang R, Shen F et al. Altered DNA Methylation Sites in Peripheral Blood Leukocytes from Patients with Simple Steatosis and Nonalcoholic Steatohepatitis (NASH). Med Sci Monit. 2018 Oct 1;24:6946-6967. doi: 10.12659/MSM.909747. PMID: 30270343; –PMCID: PMC6180948.
- Melton PE, Burton MA, Lillycrop KA et al. Differential DNA methylation of steatosis and non-alcoholic fatty liver disease in adolescence. Hepatol Int. 2023 Jun;17(3):584-594. doi: 10.1007/s12072-022-10469-7. Epub 2023 Feb 3. PMID: 36737504; PMCID: PMC9897882.
- Sharp GC, Salas LA, Monnereau C et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum Mol Genet. 2017 Oct 15;26(20):4067-4085. doi: 10.1093/hmg/ddx290. PMID: 29016858; PMCID: PMC5656174.
- Horii R, Honda M, Shirasaki T et al. MicroRNA-10a Impairs Liver Metabolism in Hepatitis C Virus-Related Cirrhosis Through Deregulation of the Circadian Clock Gene Brain and Muscle Aryl Hydrocarbon Receptor Nuclear Translocator-Like 1. Hepatol Commun. 2019 Sep 26;3(12):1687-1703. doi: 10.1002/hep4.1431. PMID: 31832575; PMCID: PMC6887665.
- Castellano-Castillo D, Moreno-Indias I, Sanchez-Alcoholado L et al. Altered Adipose Tissue DNA Methylation Status in Metabolic Syndrome: Relationships Between Global DNA Methylation and Specific Methylation at Adipogenic, Lipid Metabolism and Inflammatory Candidate Genes and Metabolic Variables. J Clin Med. 2019 Jan 13;8(1):87. doi: 10.3390/jcm8010087. PMID: 30642114; PMCID: PMC6352101.
- Kitamoto T, Kitamoto A, Ogawa Y et al. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes enco–ding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol. 2015 Aug;63(2):494-502. doi: 10.1016/j.jhep.2015.02.049. Epub 2015 Mar 14. PMID: 25776890.
- Park J, Lee DH, Ham S, Oh J, Noh JR, Lee YK, Park YJ et al. Targeted erasure of DNA methylation by TET3 drives adipogenic reprogramming and differentiation. Nat Metab. 2022 Jul;4(7):918-931. doi: 10.1038/s42255-022-00597-7. Epub 2022 Jul 4. PMID: 35788760.
- Lee JE, Schmidt H, Lai B, Ge K. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol Cell Biol. 2019 May 14;39(11):e00601-18. doi: 10.1128/MCB.00601-18. PMID: 30936246; PMCID: PMC6517598.
- Malodobra-Mazur M, Cierzniak A, Dobosz T. Oleic acid influences the adipogenesis of 3T3-L1 cells via DNA Methylation and may predispose to obesity and obesity-related disorders. Lipids Health Dis. 2019 Dec 28;18(1):230. doi: 10.1186/s12944-019-1173-6. PMID: 31883537; PMCID: PMC6935146.
- Cui TT, Huang JX, Ning BL, Mu F, Chen HY, Xing TY, Li H et al. DNA methylation promotes the expression of PPARγ transcript 1 at least in part by preventing NRF1 binding to the promoter P1 of chicken PPARγ gene. Poult Sci. 2024 Feb 16;103(5):103559. doi: 10.1016/j.psj.2024.103559. Epub ahead of print. PMID: 38430780; PMCID: PMC10912915.
- Wang Y, Chen L, Pandak WM, Heuman D, Hylemon PB, Ren S. High Glucose Induces Lipid Accumulation via 25-Hydroxycholesterol DNA-CpG Methylation. iScience. 2020 May 22;23(5):101102. doi: 10.1016/j.isci.2020.101102. Epub 2020 Apr 29. PMID: 32408171; –PMCID: PMC7225732.
- Wang J, Zhang F, Yang W, Gao D, Yang L, Yu C, Chen C, Li X, Zhang JS. FGF1 ameliorates obesity-associated hepatic steatosis by reversing IGFBP2 hypermethylation. FASEB J. 2023 Apr;37(4):e22881. doi: 10.1096/fj.202201950R. PMID: 36934380.
- Hedbacker K, Birsoy K, Wysocki RW, Asilmaz E, Ahima RS, Farooqi IS, Friedman JM. Antidiabetic effects of IGFBP2, a leptin-regulated gene. Cell Metab. 2010 Jan;11(1):11-22. doi: 10.1016/j.cmet.2009.11.007. Erratum in: Cell Metab. 2010 Mar 3;11(3):239. PMID: 20074524.
- Ding Y, Li J, Liu S, Zhang L, Xiao H, Li J, Chen H et al. DNA hypomethylation of inflammation-associated genes in adipose tissue of female mice after multigenerational high fat diet feeding. Int J Obes (Lond). 2014 Feb;38(2):198-204. doi: 10.1038/ijo.2013.98. Epub 2013 May 27. PMID: 23736364.
- Li D, Guo X, Zhao W, Jingyu J, Xia C, Yu G. Genome-wide DNA methylation dynamics in carbon tetrachloride-induced mice liver fibrosis. Iran J Basic Med Sci. 2023 Jan;26(1):85-92. doi: 10.22038/IJBMS.2022.66256.14555. PMID: 36594057; PMCID: PMC9790058.
- Götze S, Schumacher EC, Kordes C, Häussinger D. Epige–netic Changes during Hepatic Stellate Cell Activation. PLoS One. 2015 Jun 12;10(6):e0128745. doi: 10.1371/journal.pone.0128745. PMID: 26065684; PMCID: PMC4466775.;
- Caligiuri A, Gentilini A, Pastore M, Gitto S, Marra F. Cellular and Molecular Mechanisms Underlying Liver Fibrosis Regression. Cells. 2021 Oct 15;10(10):2759. doi: 10.3390/cells10102759. PMID: 34685739; PMCID: PMC8534788.
- Moran-Salvador E, Mann J. Epigenetics and Liver Fibrosis. Cell Mol Gastroenterol Hepatol. 2017 Apr 26;4(1):125-134. doi: 10.1016/j.jcmgh.2017.04.007. PMID: 28593184; PMCID: PMC5453904.
- Yang L, Liu Y, Sun Y, Huang C, Li J, Wang Y. New advances of DNA/RNA methylation modification in liver fibrosis. Cell Signal. 2022 Apr;92:110224. doi: 10.1016/j.cellsig.2021.110224. Epub 2021 Dec 24. PMID: 34954394.
- Panebianco C, Oben JA, Vinciguerra M, Pazienza V. Senescence in hepatic stellate cells as a mechanism of liver fibrosis reversal: a putative synergy between retinoic acid and PPAR-gamma signalings. Clin Exp Med. 2017 Aug;17(3):269-280. doi: 10.1007/s10238-016-0438-x. Epub 2016 Sep 21. PMID: 27655446.
- Liu R, Li Y, Zheng Q, Ding M, Zhou H, Li X. Epigenetic modification in liver fibrosis: Promising therapeutic direction with significant challenges ahead. Acta Pharm Sin B. 2024 Mar;14(3):1009-1029. doi: 10.1016/j.apsb.2023.10.023. Epub 2023 Nov 4. PMID: 38486982; –PMCID: PMC10935124.
- Zeybel M, Hardy T, Wong YK, Mathers JC, Fox CR, Gackowska A, Oakley F et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat Med. 2012 Sep;18(9):1369-77. doi: 10.1038/nm.2893. Erratum in: Nat Med. 2012 Oct;18(10):1592. PMID: 22941276; PMCID: PMC3489975.
- Hlady RA, Zhao X, Pan X, Yang JD, Ahmed F, Antwi SO, Giama NH et al. Genome-wide discovery and validation of diagnostic DNA methylation-based biomarkers for hepatocellular cancer detection in circulating cell free DNA. Theranostics. 2019 Sep 25;9(24):7239-7250. doi: 10.7150/thno.35573. PMID: 31695765; PMCID: PMC6831291.
- Zhu H, He C, Zhao H, Jiang W, Xu S, Li J, Ma T, Huang C. Sennoside A prevents liver fibrosis by binding DNMT1 and suppressing DNMT1-mediated PTEN hypermethylation in HSC activation and proliferation. FASEB J. 2020 Nov;34(11):14558-14571. doi: 10.1096/fj.202000494RR. Epub 2020 Sep 18. PMID: 32946656.
- Zeybel M, Hardy T, Robinson SM et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenetics. 2015 Mar 14;7(1):25. doi: 10.1186/s13148-015-0056-6. PMID: 25859289; –PMCID: PMC4391139.
- Maude H, Sanchez-Cabanillas C, Cebola I. Epigenetics of Hepatic Insulin Resistance. Front Endocrinol (Lausanne). 2021 May 11;12:681356. doi: 10.3389/fendo.2021.681356. PMID: 34046015; –PMCID: PMC8147868.
- PLOS ONE Staff. Correction: Branched chain amino acid transaminase 1 (BCAT1) is overexpressed and hypomethylated in patients with non-alcoholic fatty liver disease who experience adverse clinical events: A pilot study. PLoS One. 2019 Feb 5;14(2):e0212144. doi: 10.1371/journal.pone.0212144. Erratum for: PLoS One. 2018 Sep 28;13(9):e0204308. PMID: 30721255; PMCID: PMC6363212.
- Schiöth HB, Boström A, Murphy SK, Erhart W, Hampe J, Moylan C, Mwinyi J. A targeted analysis reveals relevant shifts in the methylation and transcription of genes responsible for bile acid homeostasis and drug metabolism in non-alcoholic fatty liver disease. BMC Genomics. 2016 Jun 14;17:462. doi: 10.1186/s12864-016-2814-z. PMID: 27301979; PMCID: PMC4908840.
- Sayed TS, Maayah ZH, Zeidan HA, Agouni A, Korashy HM. Insight into the physiological and pathological roles of the aryl hydrocarbon receptor pathway in glucose homeostasis, insulin resistance, and diabetes development. Cell Mol Biol Lett. 2022 Nov 22;27(1):103. doi: 10.1186/s11658-022-00397-7. PMID: 36418969; PMCID: PMC9682773.
- Wan X, Zhu X, Wang H, Feng Y, Zhou W, Liu P, Shen W et al. PGC1α protects against hepatic steatosis and insulin resistance via enhancing IL10-mediated anti-inflammatory response. FASEB J. 2020 Aug;34(8):10751-10761. doi: 10.1096/fj.201902476R. Epub 2020 Jul 7. PMID: 32633848.
- Qian L, Zhu Y, Deng C, Liang Z, Chen J, Chen Y, Wang X et al. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family in physiological and pathophysiological process and diseases. Signal Transduct Target Ther. 2024 Mar 1;9(1):50. doi: 10.1038/s41392-024-01756-w. PMID: 38424050; PMCID: PMC10904817.
- Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010 Dec;52(6):1992-2000. doi: 10.1002/hep.23927. Epub 2010 Oct 1. PMID: 20890895.
- Besse-Patin A, Jeromson S, Levesque-Damphousse P, Secco B, Laplante M, Estall JL. PGC1A regulates the IRS1:IRS2 ratio during fasting to influence hepatic metabolism downstream of insulin. Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4285-4290. doi: 10.1073/pnas.1815150116. Epub 2019 Feb 15. PMID: 30770439; PMCID: PMC6410797.
- Santos JL, Krause BJ, Cataldo LR et al. PPARGC1A Gene Promoter Methylation as a Biomarker of Insulin Secretion and Sensitivity in Response to Glucose Challenges. Nutrients. 2020 Sep 11;12(9):2790. doi: 10.3390/nu12092790. PMID: 32933059; PMCID: PMC7551463.
- Sun N, Shen C, Zhang L, Wu X, Yu Y, Yang X, Yang C et al. Hepatic Krüppel-like factor 16 (KLF16) targets PPARα to improve steatohepatitis and insulin resistance. Gut. 2021 Nov;70(11):2183-2195. doi: 10.1136/gutjnl-2020-321774. Epub 2020 Nov 30. PMID: 33257471; PMCID: PMC8515101.
- Trzaskalski NA, Vulesevic B, Nguyen MA et al. Hepatocyte-derived DPP4 regulates portal GLP-1 bioactivity, modulates glucose production, and when absent influences NAFLD progression. JCI Insight. 2023 Jan 24;8(2):e154314. doi: 10.1172/jci.insight.154314. PMID: 36472923; PMCID: PMC9977314.
- Baumeier C, Schlüter L, Saussenthaler S et al. Elevated hepatic DPP4 activity promotes insulin resistance and non-alcoholic fatty liver disease. Mol Metab. 2017 Oct;6(10):1254-1263. doi: 10.1016/j.molmet.2017.07.016. Epub 2017 Aug 4. PMID: 29031724; PMCID: PMC5641684.
- Khan RS, Bril F, Cusi K, Newsome PN. Modulation of Insulin Resistance in Nonalcoholic Fatty Liver Disease. Hepatology. 2019 Aug;70(2):711-724. doi: 10.1002/hep.30429. Epub 2019 Jul 19. PMID: 30556145.
- Barchetta I, Cimini FA, Dule S, Cavallo MG. Dipeptidyl Peptidase 4 (DPP4) as A Novel Adipokine: Role in Metabolism and Fat Homeostasis. Biomedicines. 2022 Sep 16;10(9):2306. doi: 10.3390/biomedicines10092306. PMID: 36140405; PMCID: PMC9496088.
- Wachsmuth HR, Weninger SN, Duca FA. Role of the gut-brain axis in energy and glucose metabolism. Exp Mol Med. 2022 Apr;54(4):377-392. doi: 10.1038/s12276-021-00677-w. Epub 2022 Apr 26. PMID: 35474341; PMCID: PMC9076644.
- Baumeier C, Saussenthaler S, Kammel A et al. Hepatic DPP4 DNA Methylation Associates With Fatty Liver. Diabetes. 2017 Jan;66(1):25-35. doi: 10.2337/db15-1716. Epub 2016 Oct 10. PMID: 27999105.
- Walle P, Männistö V, de Mello VD, Vaittinen M, Perfilyev A, Hanhineva K, Ling C et al. Liver DNA methylation of FADS2 associates with FADS2 genotype. Clin Epigenetics. 2019 Jan 17;11(1):10. doi: 10.1186/s13148-019-0609-1. Erratum in: Clin Epigenetics. 2019 Mar 12;11(1):47. PMID: 30654845; PMCID: PMC6337806.
- Walle P, Takkunen M, Männistö V, Vaittinen M, Lankinen M, Kärjä V, Käkelä P et al. Fatty acid metabolism is altered in non-alcoholic steatohepatitis independent of obesity. Metabolism. 2016 May;65(5):655-666. doi: 10.1016/j.metabol.2016.01.011. Epub 2016 Jan 23. PMID: 27085774.
- Bláhová Z, Harvey TN, Pšenička M, Mráz J. Assessment of Fatty Acid Desaturase (Fads2) Structure-Function Properties in Fish in the Context of Environmental Adaptations and as a Target for Genetic Engineering. Biomolecules. 2020 Jan 31;10(2):206. doi: 10.3390/biom10020206. PMID: 32023831; PMCID: PMC7072455.
- Shetty SS, Suchetha KN, Harshini D, Sharmila KP, Rai S. Association of FADS2 rs174575 gene polymorphism and insulin resistance in type 2 diabetes mellitus. Afr Health Sci. 2020 Dec;20(4):1770-1776. doi: 10.4314/ahs.v20i4.30. PMID: 34394238; PMCID: PMC8351823.
- Shetty SS, Kumari NS. Fatty acid desaturase 2 (FADS 2) rs174575 (C/G) polymorphism, circulating lipid levels and susceptibility to type-2 diabetes mellitus. Sci Rep. 2021 Jun 23;11(1):13151. doi: 10.1038/s41598-021-92572-7. PMID: 34162950; PMCID: PMC8222307.
- Qiu YY, Zhang J, Zeng FY, Zhu YZ. Roles of the peroxisome proliferator-activated receptors (PPARs) in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). Pharmacol Res. 2023 Jun;192:106786. doi: 10.1016/j.phrs.2023.106786. Epub 2023 May 3. PMID: 37146924.
- Jonas W, Schürmann A. Genetic and epigenetic factors determining NAFLD risk. Mol Metab. 2021 Aug; 50:101111. doi: 10.1016/j.molmet.2020.101111. Epub 2020 Nov 5. PMID: 33160101; PMCID: PMC8324682.